
Autism spectrum disorder (ASD) is the most common neurodevelopmental disorder with a constantly increasing prevalence. A simple animal such as the nematode Caenorhabditis elegans may provide insights into the extreme complexity of ASD genetics. In this study, we present a screening approach of potential C. elegans mutants as potential ASD models. We screened these mutants for motor-deficiency phenotypes, which can be exploited to study underlying mechanisms of the disorder. Selected motordeficient mutants were then used in a comprehensive drug screen of over 3900 compounds, including many FDA-approved and natural molecules, that were analyzed for their ability to suppress motility defects caused by ASD-associated gene orthologues.
Materials And Methods:
Fort the discovery of novel potential ASD C. elegans models, mutants strains were screened for motor-deficiency phenotypes using a worm tracking machine (Wmicrotraker) in 96-well-microtiter plates. Approximately 30 worms per well were put in M9 buffer and their swimming movements were tracked for at least 6 hours. Movement of each strain was measured in triplicates.
Once the potential models for ASD were identified, the compounds were tested to evaluate the reversion of the defective phenotype. The motor deficient worm strains were exposed to the drugs in the microtiter wells at a concentration of 20 µM in DMSO while swimming in M9 buffer with OP50 and movement was tracked for at least 4 hours. Measurements were performed in triplicates and the average movement score was compared to a no-compound control and the average movement score of the whole plate. If the values for a certain drug were higher than the respective controls, a second screen was performed to validate the increased movement due to the specific drug. Candidates that increased movement abilities significantly in the second screen were counted as positive.
Results:
From the 27 genes involved in ASD that were tested via screening of 37 mutant strains, 12 gene mutations in 16 mutant strains cause significantly impaired swimming locomotion : nrx-1(ok1649), mbk-1(ok402), nmr-2(ok3324), fmi-1(hd121), nlg-1(ok259), chd7(gk290), egl-19(n582), dys-1(eg33), unc-2(ra612), unc-2(e55), unc-2(gk366), fmi-1(rh308), mca-3(ar492), unc-6(e78), mca-3(ar493), and unc-44(e362). Drug screen of compounds that rescue motor-deficient phenotypes of the mutant strains that were identified with impaired swimming locomotion; we chose four strains to perform a comprehensive drug screen that exploited their motor deficient phenotypes: unc-44(e362), egl-19(n582), nmr-2(ok3324), and unc-2(ra612).
The movement of these four strains is shown in Figure 2. From the best 4 mutant strains, model of autism, positively acting compounds that corrected the impaired swimming movement phenotypes of each strain are listed. 49, 56, 62 and 28 substances were found to have beneficial effects in unc-44(e362), egl-19(n582), nmr- 2(ok3324), and unc-2(ra612), respectively.
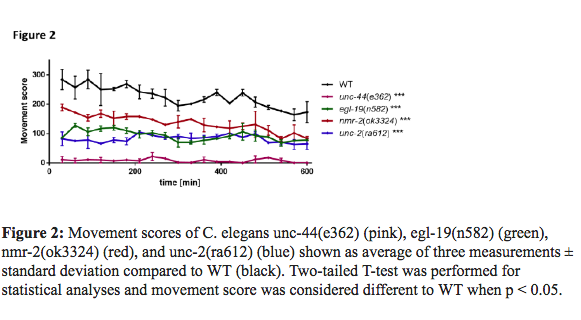
…………………………………………………………………………………………………………………………..
Exp Neurol. 2017 Jul;293:101-114. doi: 10.1016/j.expneurol.2017.03.022. Epub 2017 Apr 1.
Schmeisser K, Fardghassemi Y, Parker JA.
